No one had ever seen one virus latching onto another virus, until anomalous sequencing results sent a UMBC team down a rabbit hole leading to a first-of-its-kind discovery.
It’s known that some viruses, called satellites, depend not only on their host organism to complete their life cycle, but also on another virus, known as a “helper,” explains Ivan Erill, professor of biological sciences. The satellite virus needs the helper either to build its capsid, a protective shell that encloses the virus’s genetic material, or to help it replicate its DNA. These viral relationships require the satellite and the helper to be in proximity to each other at least temporarily, but there were no known cases of a satellite actually attaching itself to a helper—until now.
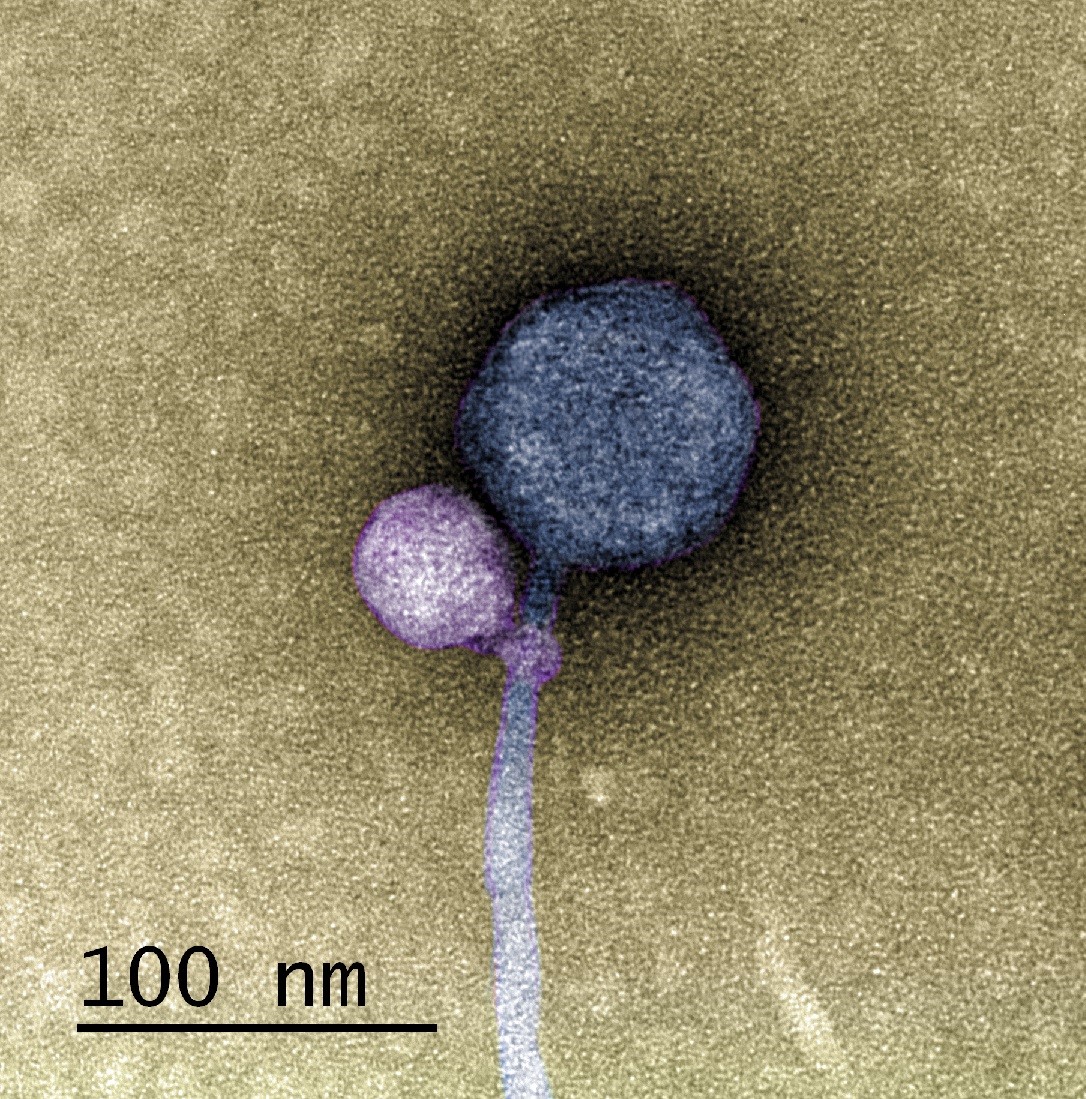
In a paper published in the Journal of the International Society of Microbial Ecology, a UMBC team and colleagues from Washington University in St. Louis (WashU) describe the first observation of a satellite bacteriophage (a virus that infects bacterial cells) consistently attaching to a helper bacteriophage at its “neck”—where the capsid joins the tail of the virus.
In detailed electron microscopy images taken by Tagide deCarvalho, assistant director of the College of Natural and Mathematical Sciences core facilities and first author on the new paper, 80 percent (40 out of 50) helpers had a satellite bound at the neck. Some of those that did not had remnant satellite tendrils present at the neck. Erill, senior author on the paper, describes them as appearing like “bite marks.”
“When I saw it, I was like, ‘I can’t believe this,’” deCarvalho says. “No one has ever seen a bacteriophage—or any other virus—attach to another virus.”

A long-term virus relationship
After the initial observations, Elia Mascolo, a graduate student in Erill‘s research group and co-first author on the paper, analyzed the genomes of the satellite, helper, and host, which revealed further clues about this never-before-seen viral relationship. Most satellite viruses contain a gene that allows them to integrate into the host cell’s genetic material after they enter the cell. This allows the satellite to reproduce whenever a helper happens to enter the cell from then on. The host cell also copies the satellite’s DNA along with its own when it divides.

A bacteriophage sample from WashU also contained a helper and a satellite. The WashU satellite has a gene for integration and does not directly attach to its helper, similar to previously observed satellite-helper systems.
However, the satellite in UMBC’s sample, named MiniFlayer by the students who isolated it, is the first known case of a satellite with no gene for integration. Because it can’t integrate into the host cell’s DNA, it must be near its helper—named MindFlayer—every time it enters a host cell if it is going to survive. Given that, although the team did not directly prove this explanation, “Attaching now made total sense,” Erill says, “because otherwise, how are you going to guarantee that you are going to enter into the cell at the same time?”
Additional bioinformatics analysis by Mascolo and Julia López-Pérez, another Ph.D. student working with Erill, revealed that MindFlayer and MiniFlayer have been co-evolving for a long time. “This satellite has been tuning in and optimizing its genome to be associated with the helper for, I would say, at least 100 million years,” Erill says, which suggests there may be many more cases of this kind of relationship waiting to be discovered.
Contamination or discovery?
This groundbreaking discovery could easily have been missed. The project started out as a typical semester in the SEA-PHAGES program—an investigative curriculum where undergraduates isolate bacteriophages from environmental samples, send them out for sequencing, and then use bioinformatics tools to analyze the results. When the sequencing lab at the University of Pittsburgh reported contamination in the sample from UMBC expected to contain the MindFlayer phage, the journey began.

The sample included one large sequence: the phage they expected. “But instead of just finding that, we also found a small sequence, which didn’t map to anything we knew,” says Erill, who is also one of the leads for UMBC’s SEA-PHAGES program, called Phage Hunters, along with Steven Caruso, principal lecturer of biological sciences. Caruso ’94, Ph.D. ’02, biological sciences, ran the isolation again, sent it out for sequencing—and got identical results.
That’s when the team pulled in deCarvalho to get a visual of what was going on with the transmission electron microscope (TEM) at UMBC’s Keith R. Porter Imaging Facility (KPIF). Without the images, the discovery would have been impossible.
“Not everyone has a TEM at their disposal,” deCarvalho notes. But with the instruments at the KPIF, deCarvalho says, “I’m able to follow up on some of these observations and validate them with imaging. There’s elements of discovery we can only make using the TEM.”
The team’s discovery sets the stage for future work to figure out how the satellite attaches, how common this phenomenon is, and much more. “It’s possible that a lot of the bacteriophages that people thought were contaminated were actually these satellite-helper systems,” deCarvalho says. “So now, with this paper, people might be able to recognize more of these systems.”